On August 29th, a collaborative research team including Professor Zheng Ke and Professor Guo Xuejiang from the State Key Laboratory of Reproductive Medicine and Offspring Health at NMU, Associate Professor Lin Mingyan from the Department of Neurobiology at NMU’s School of Basic Medical Sciences, and Professor Tan Yueqiu from Central South University, published a research article titled “The Landscape of RNA-Binding Proteins in Mammalian Spermatogenesis” online in the academic journal Science.

The mammalian testes transcribe a large portion of the genome, making them one of the organs with the most complex tissue transcriptomes. However, there’s a lack of systematic understanding of how RNA-binding proteins (RBPs) dynamically regulate the extensive array of RNAs during spermatogenesis. In particular, nondomain elements (short motifs not included in annotated domain) of RBPs have garnered growing research interest but remain underexplored in animal models. Despite the link between individual RBP dysfunction and various diseases, there is a lack of comprehensive genome-wide insights into how these deficiencies correlate with specific conditions, including male infertility, a significant global reproductive issue.
To systematically identify RBPs in mouse male germ cells (mMGCs) at different stages, the team isolated spermatogonia, pachytene spermatocytes, and round spermatids and performed RNA interactome capture. The team established a comprehensive atlas of mMGC RBPs, including those with previously unknown RNA-related activities specialized for meiotic and post-meiotic functions. The article showed that mMGC RBPs were involved in multi-layered dynamics covering proteomic expression patterns, proteo-transcriptomic discordancy, relative RNA-binding activity, and ribonucleoprotein (RNP) concordant behavior.
Next, the team employed RBDmap method to capture RNA-crosslinked peptides from testes, enabling the screening of RNA-binding domains and nondomain elements, followed by bioinformatic and experimental characterization of their prevalence, property, and function using orthogonal approaches.The team discovered a polyampholytic nondomain element known as glutamic acid-arginine (ER) patch, a widely existing short fragment rich in E and R. This motif is positioned adjacent to the coiled-coil (CC) region of RBP, shows evolutionary co-variation of nearby residue pairs, and significantly enhances RNA binding of its host RBPs.
The team focused on the ER patch in RNA-binding protein NONO (non-POU domain-containing octamer-binding protein) and used animal models to demonstrate its role in regulating the transition of spermatogenic cells from mitosis to meiosis at the single-cell transcriptome level.
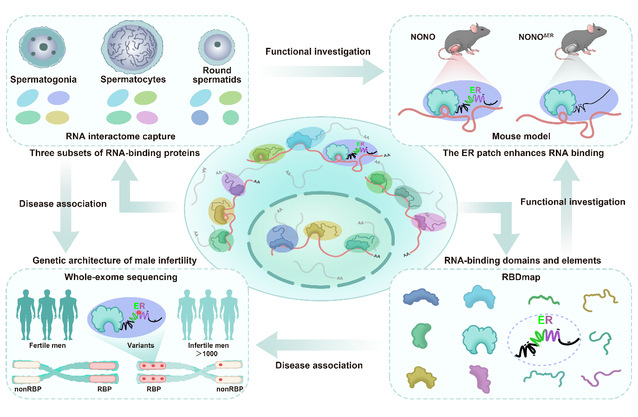
The team also integrated large whole-exome sequencing profiles from male infertile cohorts with non-obstructive azoospermia or oligozoospermia. This approach revealed the prominent role of RNA-binding proteins in male infertility and highlighted the potential clinical pathogenicity of nondomain elements, such as ER patches at the systemic level.
This study provides a comprehensive and rich resource for RNA-binding proteins related to spermatogenesis and male infertility. It underscores the functional significance of the nondomain element in regulating RNA binding and spermatogenesis, highlighting the extensive value of this resource for decoding the genetic and molecular foundations of male fertility.This achievement, published in Science and held under NMU’s fully independent intellectual property, marks a significant breakthrough for our reproductive medicine research team in the field of male reproduction.
(Drafted by Zheng Ke Research Group; Reviewed by the State Key Laboratory of Reproductive Medicine and Offspring Health and Research Institute; Translation revised by Zhang Bei)



